Making eDNA FAIR
Why do we need FAIR eDNA?
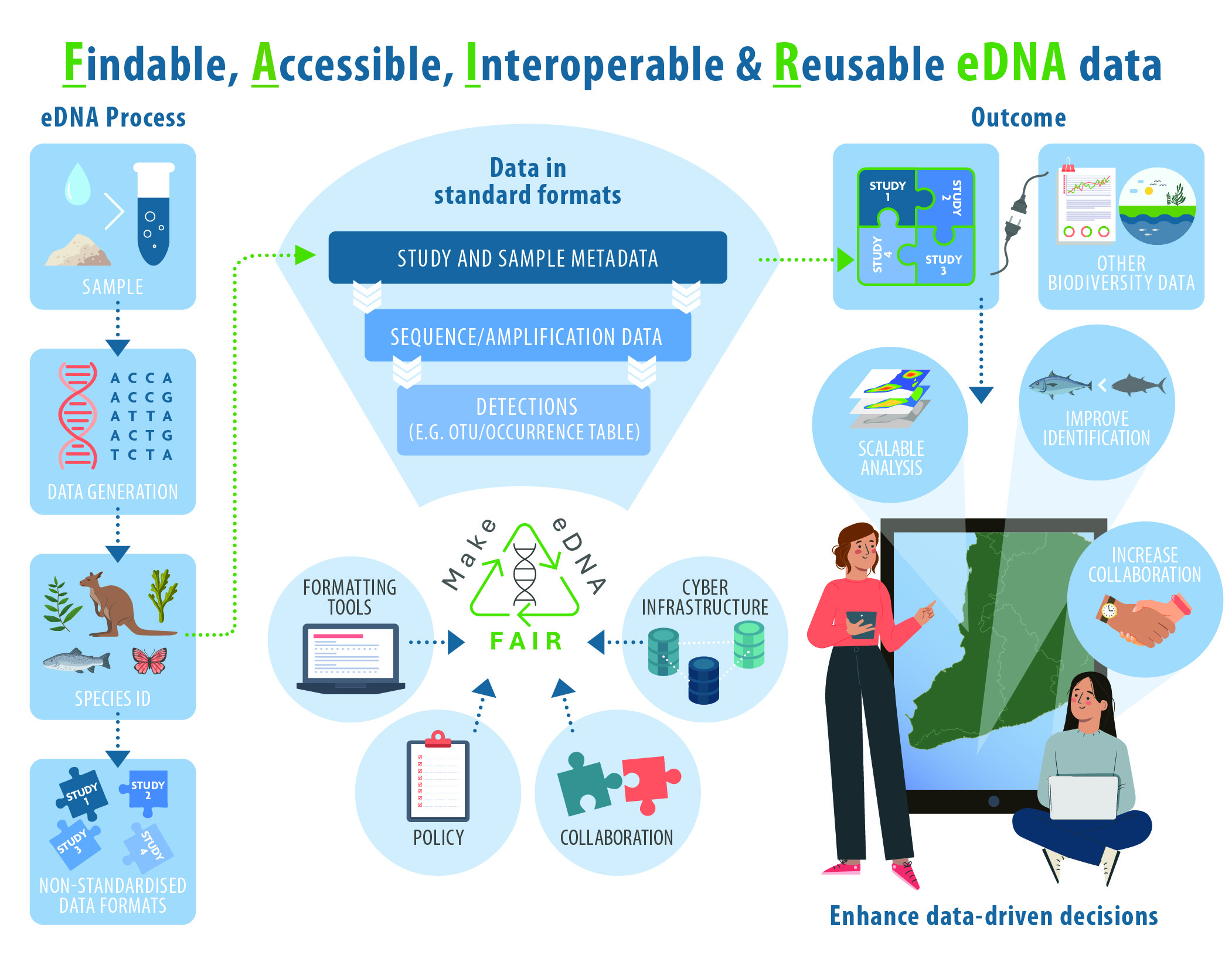
The success of environmental DNA (eDNA) approaches for species detection has revolutionised biodiversity monitoring and distribution mapping. Targeted eDNA amplification approaches, such as quantitative PCR, have improved our understanding of species distribution, while metabarcoding-based approaches enable biodiversity assessment at unprecedented scales and taxonomic resolution. eDNA datasets, however, are often scattered across repositories with inconsistent formats, varying access restrictions, and inadequate metadata; this limits their interoperation, reuse, and overall impact. Adopting FAIR (Findable, Accessible, Interoperable, Reusable) data practices with eDNA data can transform monitoring of biodiversity and individual species, and support data-driven biodiversity management across broad scales. FAIR practices remain underdeveloped in the eDNA community, partly due to gaps in adapting existing vocabularies, such as Darwin Core (DwC) and Minimum Information about any (x) Sequence (MIxS), to eDNA-specific needs and workflows.
Project scope
Fine-tuning, optimizing and extending existing data and metadata standards specifically for eDNA-based studies is a crucial first step towards achieving FAIR eDNA. To this end, we propose a comprehensive FAIR eDNA (FAIRe) metadata checklist, which integrates existing data standards and introduces new terms tailored to eDNA workflows. Metadata are systematically linked to both raw data (e.g., metabarcoding sequences, Ct/Cq values of targeted qPCR assays) and derived biological observations (e.g., ASV/OTU tables, species presence/absence). Along with formatting guidelines, tools, templates and example datasets, we introduce a standardised, ready-to-use approach for FAIR eDNA practices. Together, these efforts aim to guide data providers and facilitate the unambiguous description of eDNA datasets that are aligned with FAIR data practices. Our goal is to provide a FAIRe metadata checklist and guidelines as resources for existing data standards, infrastructures and databases, helping them to address gaps to better meet their own objectives. The guidelines will also benefit individual eDNA science practitioners and labs by improving data management standardisation, making their data more FAIR not only for reusers, but also for internal organisation.